Toy Example - Deep prior#
import tensorflow_probability as tfp
import matplotlib.pyplot as plt
import tensorflow as tf
import seaborn as sns
import pandas as pd
import numpy as np
import elicit as el
from bayesflow.inference_networks import InvertibleNetwork
from elicit.extras import utils
tfd = tfp.distributions
The Model#
Generative model#
Implementation#
Predictor#
# create a predictor ranging from 1 to 200
# standardize predictor
# select the 5th, 25th, 50th, 75th, and 95th quantile of the std. predictor for querying the expert
def std_predictor(N, quantiles):
X = tf.cast(np.arange(N), tf.float32)
X_std = (X-tf.reduce_mean(X))/tf.math.reduce_std(X)
X_sel = tfp.stats.percentile(X_std, quantiles)
return X_sel
std_predictor(N=200, quantiles=[5,25,50,75,95])
<tf.Tensor: shape=(5,), dtype=float32, numpy=
array([-1.5502049 , -0.85737586, 0.00866036, 0.85737586, 1.5502049 ],
dtype=float32)>
Generative model#
class ToyModel:
def __call__(self, prior_samples, design_matrix, **kwargs):
B = prior_samples.shape[0]
S = prior_samples.shape[1]
# preprocess shape of design matrix
X = tf.broadcast_to(design_matrix[None, None,:],
(B,S,len(design_matrix)))
# linear predictor (= mu)
epred = tf.add(prior_samples[:, :, 0][:,:,None],
tf.multiply(prior_samples[:, :, 1][:,:,None], X)
)
# data-generating model
likelihood = tfd.Normal(
loc=epred, scale=tf.expand_dims(prior_samples[:, :, -1], -1)
)
# prior predictive distribution (=height)
ypred = likelihood.sample()
# selected observations
y_X0, y_X1, y_X2, y_X3, y_X4 = (ypred[:,:,0], ypred[:,:,1], ypred[:,:,2],
ypred[:,:,3], ypred[:,:,4])
# log R2 (log for numerical stability)
log_R2 = utils.log_R2(ypred, epred)
# correlation between parameters
cor = utils.pearson_correlation(prior_samples)
return dict(
likelihood=likelihood,
ypred=ypred, epred=epred,
prior_samples=prior_samples,
y_X0=y_X0, y_X1=y_X1, y_X2=y_X2,
y_X3=y_X3, y_X4=y_X4,
log_R2=log_R2,
cor=cor
)
Model input for elicit method#
# specify the model
model=el.model(
obj=ToyModel,
design_matrix=std_predictor(N=200, quantiles=[5,25,50,75,95])
)
Model parameters#
intercept parameter \(\beta_0\)
slope parameter \(\beta_1\)
error term \(\sigma\)
To be learned hyperparameters \(\lambda\): reflecting the weights of the deep neural networks within the normalizing flow architecture.
Parameter input for elicit method#
parameters=[
el.parameter(name="beta0"),
el.parameter(name="beta1"),
el.parameter(name="sigma")
]
Target quantities and elicitation techniques#
Target quantities
query expert regarding prior predictions \(y \mid X_{i}\) with \(i\) being the 5th, 25th, 50th, 75th, and 95th quantile of the predictor.
\(R^2 = \frac{\text{Var}(\mu)}{\text{Var}(y)}\) (we use \(\log R^2\) for numerical stability)
correlation between model parameters
Elicitation technique
query each prior prediction using quantile-based elicitation using \(Q_p(y \mid X)\) for \(p=5, 25, 50, 75, 95\)
query \(R^2\) using quantile-based elicitation using \(Q_p(y \mid X)\) for \(p=5, 25, 50, 75, 95\)
regarding the correlation structure, we assume independence between the model parameters (thus, \(\boldsymbol{\rho}_\boldsymbol{\theta}=\mathbf{0}\))
Importance of elicited statistics in loss
all elicited statistics should have equal importance (weight=1.0)
for computing the discrepancy between expert-elicited statistics and model simulations with use the Maximum Mean Discrepancy with Energy kernel
Targets input for elicit method#
targets=[
el.target(
name=f"y_X{i}",
query=el.queries.quantiles((5, 25, 50, 75, 95)),
loss=el.losses.MMD2(kernel="energy"),
weight=1.0
) for i in range(5)
]+[
el.target(
name="log_R2",
query=el.queries.quantiles((5, 25, 50, 75, 95)),
loss=el.losses.MMD2(kernel="energy"),
weight=1.0
),
el.target(
name="cor",
query=el.queries.identity(),
loss=el.losses.L2,
weight=1.0
)
]
Expert elicitation#
instead of querying a “real” expert, we define a ground truth (i.e., oracle) and simulate the oracle-elicited statistics
Expert input for elicit method (here: oracle)#
# specify ground truth
ground_truth = {
"beta0": tfd.Normal(loc=5, scale=1),
"beta1": tfd.Normal(loc=2, scale=1),
"sigma": tfd.HalfNormal(scale=10.0),
}
# define oracle
expert=el.expert.simulator(
ground_truth = ground_truth,
num_samples = 10_000
)
Normalizing Flow#
ToDo
network=el.networks.NF(
inference_network=InvertibleNetwork,
network_specs=dict(
num_params=3,
num_coupling_layers=3,
coupling_design="affine",
coupling_settings={
"dropout": False,
"dense_args": {
"units": 128,
"activation": "relu",
"kernel_regularizer": None,
},
"num_dense": 2,
},
permutation="fixed"
),
base_distribution=el.networks.base_normal
)
Training: Learn prior distributions based on expert data#
All inputs for elicit method
elicit = el.Elicit(
model=model,
parameters=parameters,
targets=targets,
expert=expert,
optimizer=el.optimizer(
optimizer=tf.keras.optimizers.Adam,
learning_rate=0.001,
clipnorm=1.0
),
trainer=el.trainer(
method="deep_prior",
name="toy",
seed=3,
epochs=200
),
network=network
)
# run method
hist = elicit.fit(save_dir=None)
Training
100%|██████████| 200/200 [05:40<00:00, 1.70s/it]
Results#
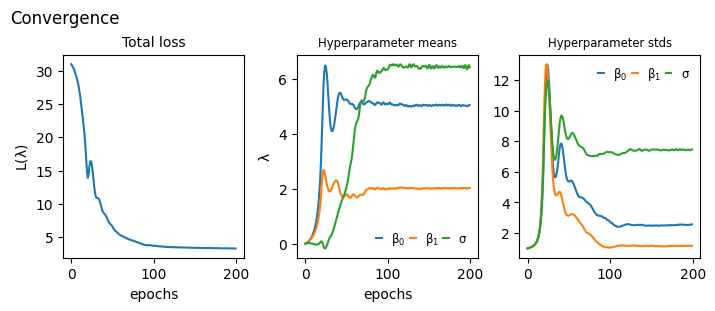
Convergence#
def plot_convergence(hist):
_, axs = plt.subplots(1,3, constrained_layout=True, figsize=(7,3), sharex=True)
axs[0].plot(hist["loss"])
for i,k in enumerate(hist["hyperparameter"]):
axs[i+1].plot(hist["hyperparameter"][k], label=[r"$\beta_0$", r"$\beta_1$", r"$\sigma$"])
axs[i+1].legend(handlelength=0.5, fontsize="small", frameon=False, ncol=3, columnspacing=0.4)
axs[i+1].set_title(f"Hyperparameter {k}", size="small")
axs[0].set_title("Total loss", size="medium")
axs[0].set_ylabel(r"$\mathcal{L}(\lambda)$")
axs[1].set_ylabel(r"$\lambda$")
[axs[i].set_xlabel("epochs") for i in range(2)]
plt.suptitle("Convergence", ha="left", x=0)
plt.show()
plot_convergence(hist)
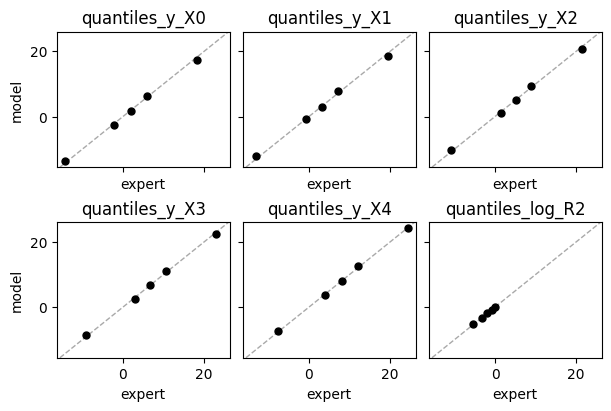
Expert expectations#
def plot_expert_data(res):
_, axs = plt.subplots(2,3, constrained_layout=True, figsize=(6,4), sharex=True, sharey=True)
for j,key in enumerate(res["expert_elicited_statistics"]):
if key=="identity_cor":
continue
if j<3:
axs[0,j].axline((0,0), slope=1, color="darkgrey", linestyle="dashed", lw=1)
axs[0,j].plot(res["expert_elicited_statistics"][key][0,:],
tf.reduce_mean(res["elicited_statistics"][key],0), "o", ms=5, color="black")
axs[0,j].set_title(key)
axs[0,j].set_xlabel("expert")
else:
axs[1,j-3].axline((0,0), slope=1, color="darkgrey", linestyle="dashed", lw=1)
axs[1,j-3].plot(res["expert_elicited_statistics"][key][0,:],
tf.reduce_mean(res["elicited_statistics"][key],0), "o", ms=5, color="black")
axs[1,j-3].set_title(key)
axs[1,j-3].set_xlabel("expert")
[axs[i,0].set_ylabel("model") for i in range(2)]
plt.show()
plot_expert_data(elicit.results)
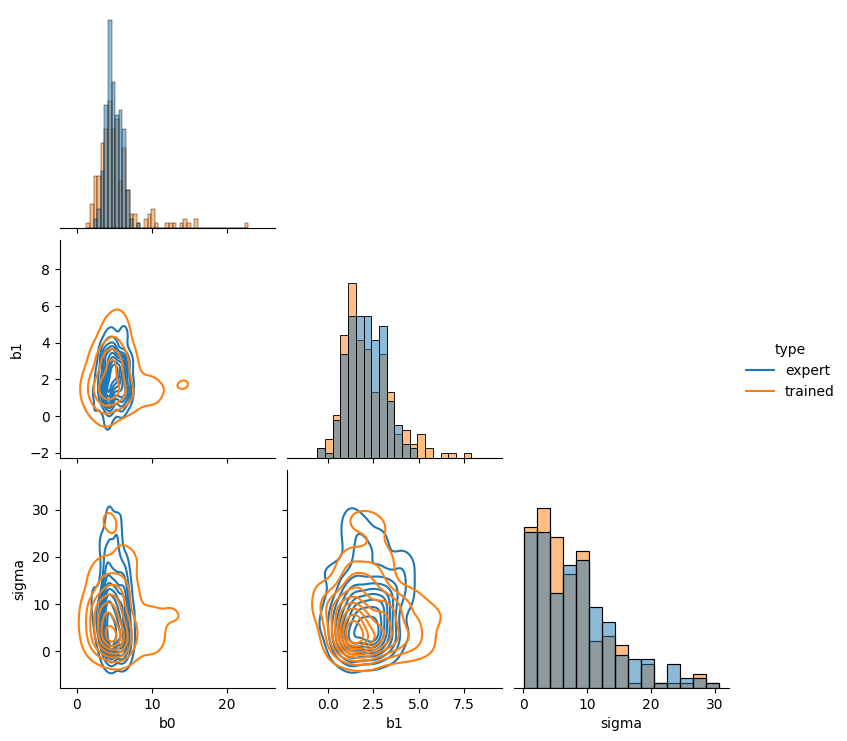
Learned joint prior#
# extract number of samples from simulated priors
num_samples = elicit.trainer["num_samples"]
# create pandas dataframes out of trained priors and orcale priors
model_priors_df = pd.DataFrame(elicit.results["prior_samples"][0,:,:],
columns = ["b0", "b1", "sigma"])
oracle_priors_df = pd.DataFrame(elicit.results["expert_prior_samples"][0,:num_samples,:],
columns = ["b0", "b1", "sigma"])
# add label to sub-dataframes
model_priors_df["type"]="trained"
oracle_priors_df["type"]="true"
# concat dataframes and ensure sigma is pos.
df = pd.concat([expert_priors_df, model_priors_df])
df["sigma"] = abs(df["sigma"])
# plot joint prior distribution
sns.pairplot(df, diag_kind="hist", hue="type", kind="kde", corner=True)
plt.show()
Add-on: Model averaging#
ToDo